Lecture 9
SparkML
Georgetown University
Fall 2023
Logistics and Review
Deadlines
Lab 7: Spark DataFrames Due Oct 10 6pmLab 8: SparkNLP Due Oct 17 6pm- Assignment 6: Spark (Multi-part) Due Oct 23 11:59pm
- Lab 9: SparkML Due Oct 24 6pm
- Lab 10: Spark Streaming Due Oct 31 6pm
- Lab 11: Dask Due Nov 7 6pm
- Lab 12: Ray Due Nov 14 6pm
Look back and ahead
- Searching Slack for existing Q&A - like StackOverflow!
- Spark RDDs, Spark DataFrames, Spark UDFs
- Now: SparkML and project
- Next week: Spark Streaming
Review of Prior Topics
AWS Academy
Credit limit - $100
Course numbers:
- Course #1 - 54380
- Course #2 - 54378
STAY WITH COURSE 54380 UNLESS YOU HAVE RUN OUT OF CREDITS OR >$90 USED!
Note that you will have to repeat several setup steps:
- SageMaker domain set up (review Slack channel to review solutiosn to issues)
- any S3 uploading or copying as well as bucket creation as necessary
Caching and Persistence
By default, RDDs are recomputed every time you run an action on them. This can be expensive (in time) if you need to use a dataset more than once.
Spark allows you to control what is cached in memory.
To tell spark to cache an object in memory, use persist() or cache():
cache():is a shortcut for using default storage level, which is memory onlypersist():can be customized to other ways to persist data (including both memory and/or disk)
collect CAUTION

Assumption: knowledge of ML techniques and process
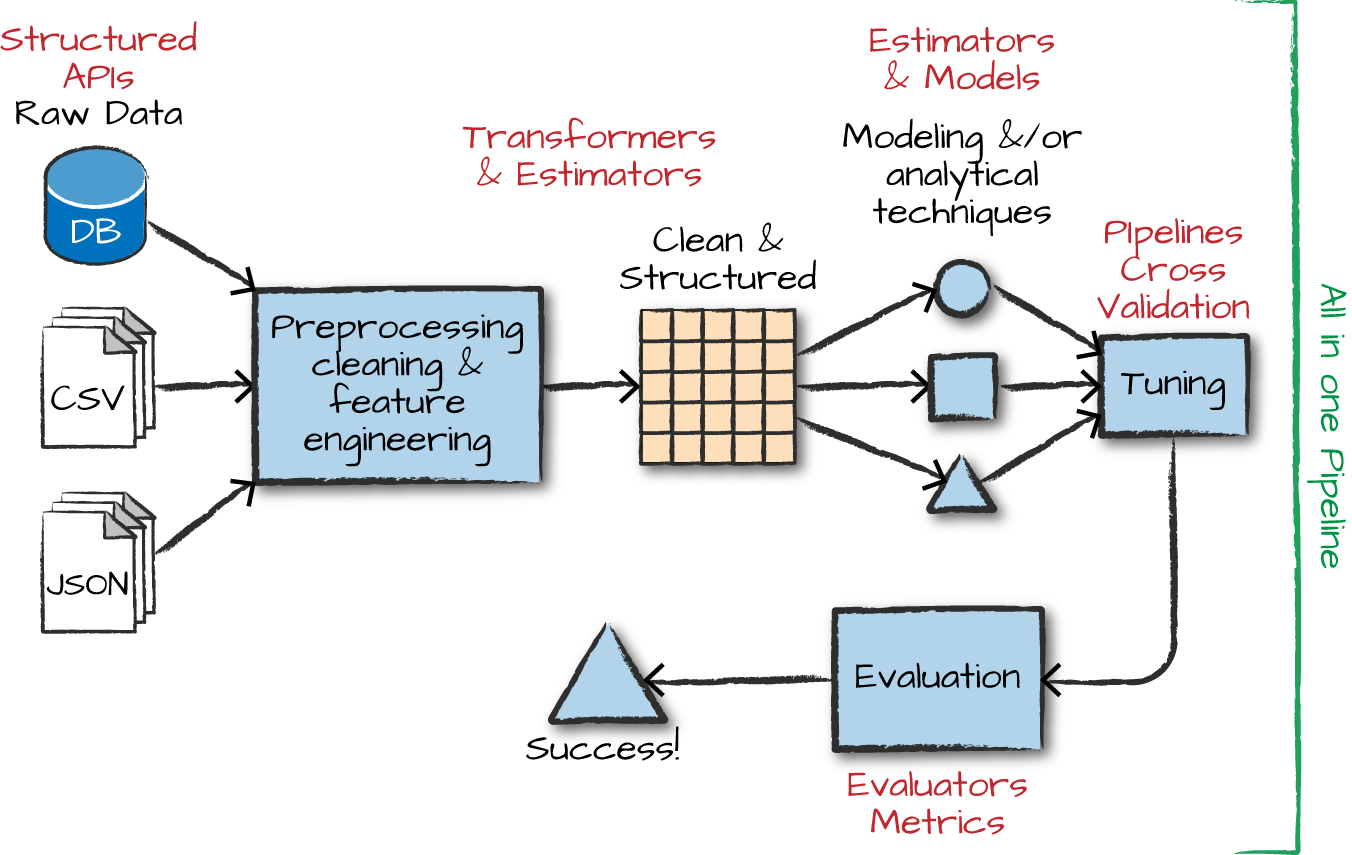
Overview of ML
The advanced analytics/ML process
Gather and collect data
Clean and inspect data
Perform feature engineering
Split data into train/test
Evaluate and compare models
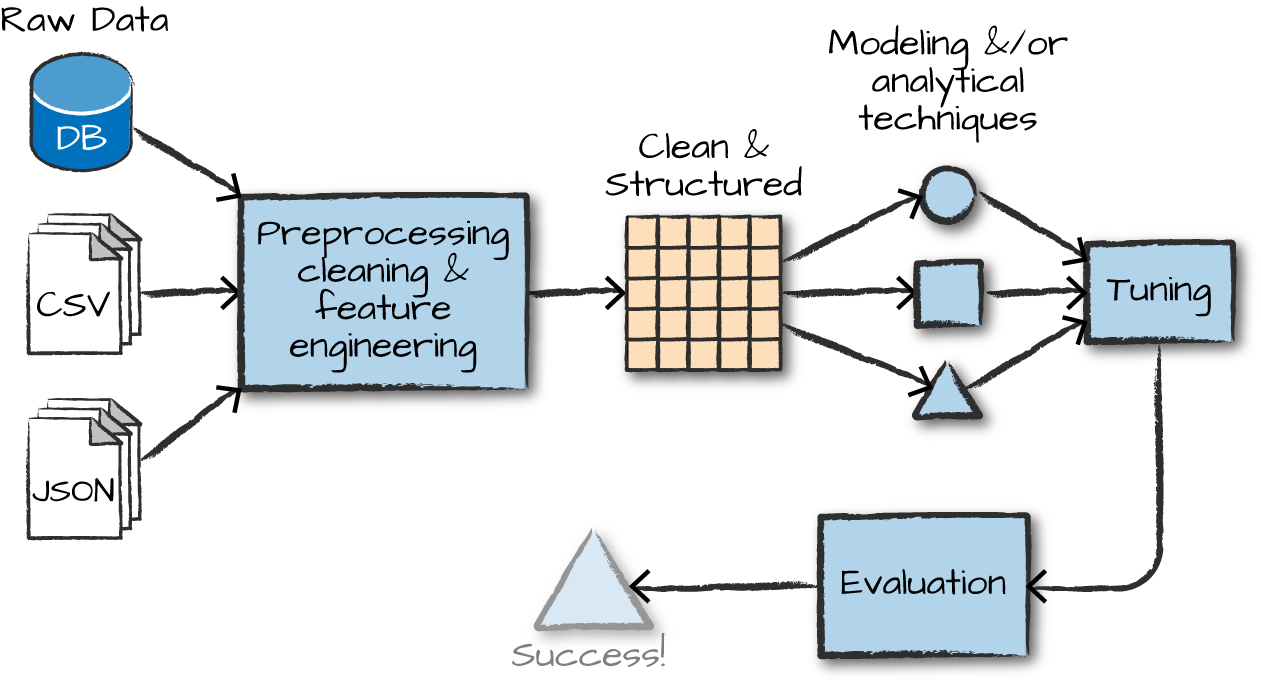
ML with Spark
What is MLlib
Capabilities
- Gather and clean data
- Perform feature engineering
- Perform feature selection
- Train and tune models
- Put models in production
API is divided into two packages
org.apache.spark.ml (High-level API) - Provides higher level API built on top of DataFrames - Allows the construction of ML pipelines
org.apache.spark.mllib (Predates DataFrames - in maintenance mode) - Original API built on top of RDDs
Inspired by scikit-learn
DataFrameTransformerEstimatorPipelineParameter
Transformers, estimators, and pipelines
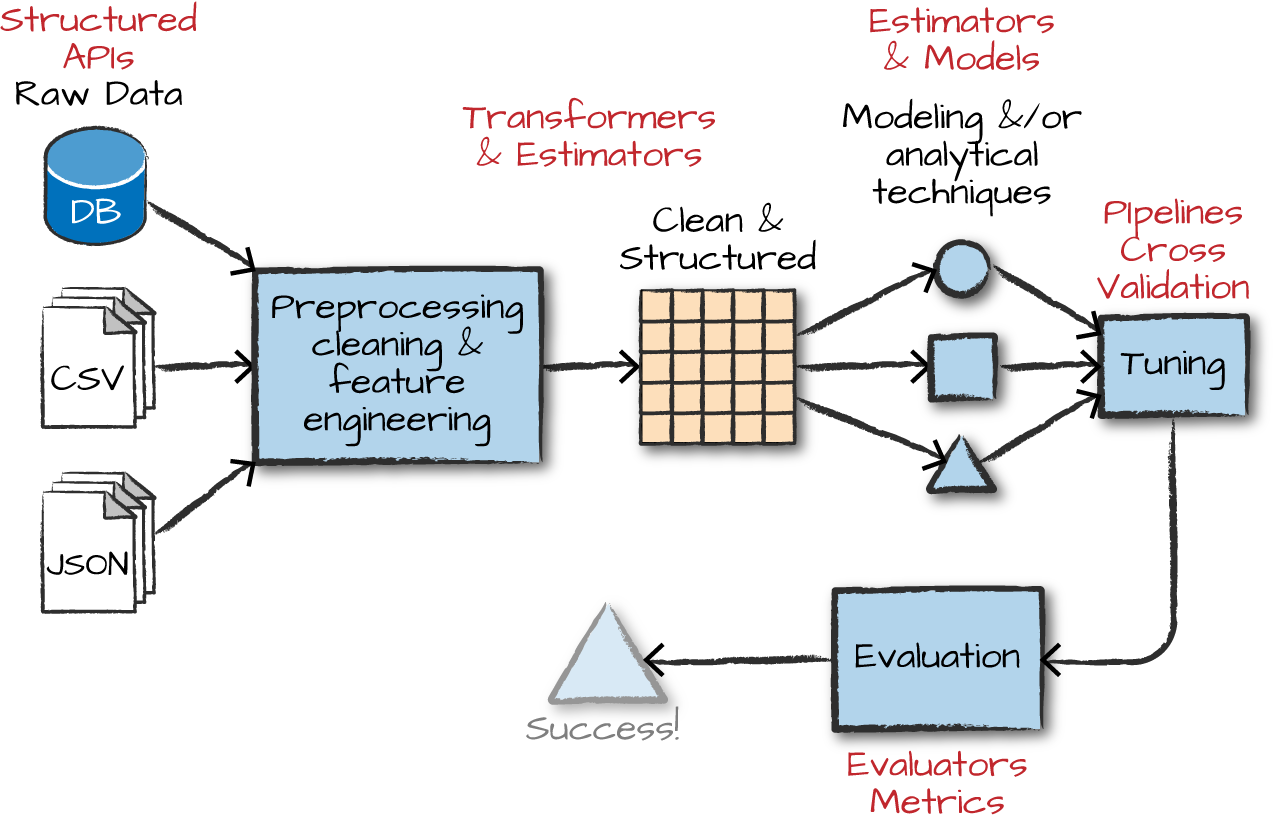
Transformers
Transformers takes DataFrames as input, and return a new DataFrame as output. Transformers do not learn any parameters from the data, they simply apply rule-based transformations to either prepare data for model training or generate predictions using a trained model.
Transformers are run using the .transform() method
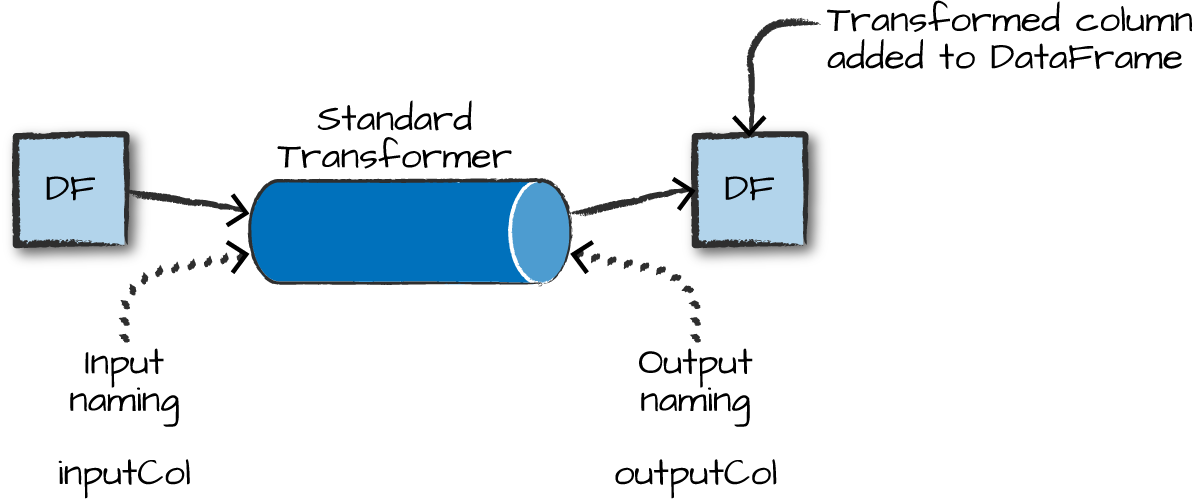
Some Examples of Transformers
- Converting categorical variables to numeric (must do this)
StringIndexerOneHotEncodercan act on multiple columns at a time
- Data Normalization
NormalizerStandardScaler
- String Operations
TokenizerStopWordsRemoverPCA
- Converting continuous to discrete
BucketizerQuantileDiscretizer
- Many more
- https://spark.apache.org/docs/latest/ml-guide.html
MLlib algorithms for machine learning models need single, numeric features column as input
Each row in this column contains a vector of data points corresponding to the set of features used for prediction.
Use the
VectorAssemblertransformer to create a single vector column from a list of columns.All categorical data needs to be numeric for machine learning
# Example from Spark docs
from pyspark.ml.linalg import Vectors
from pyspark.ml.feature import VectorAssembler
dataset = spark.createDataFrame(
[(0, 18, 1.0, Vectors.dense([0.0, 10.0, 0.5]), 1.0)],
["id", "hour", "mobile", "userFeatures", "clicked"])
assembler = VectorAssembler(
inputCols=["hour", "mobile", "userFeatures"],
outputCol="features")
output = assembler.transform(dataset)
print("Assembled columns 'hour', 'mobile', 'userFeatures' to vector column 'features'")
output.select("features", "clicked").show(truncate=False)Estimators
Estimators learn (or “fit”) parameters from your DataFrame via the .fit() method, and return a model which is a Transformer
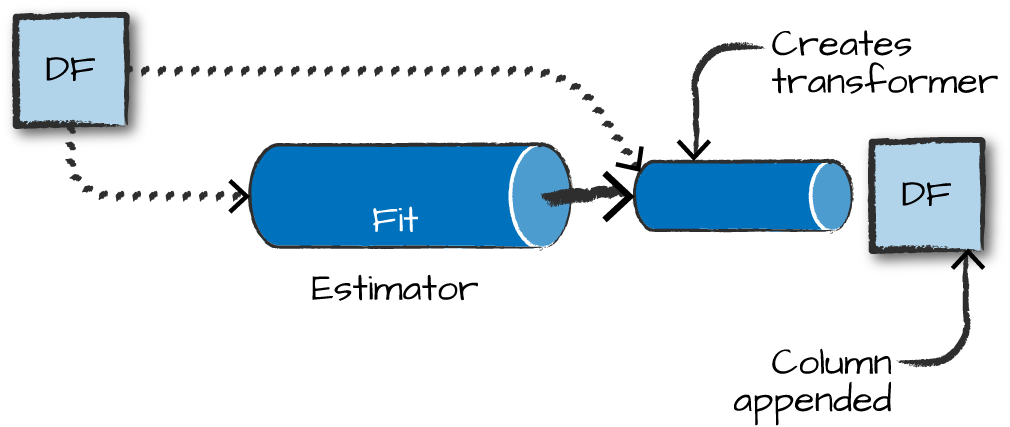
Pipelines
Pipelines
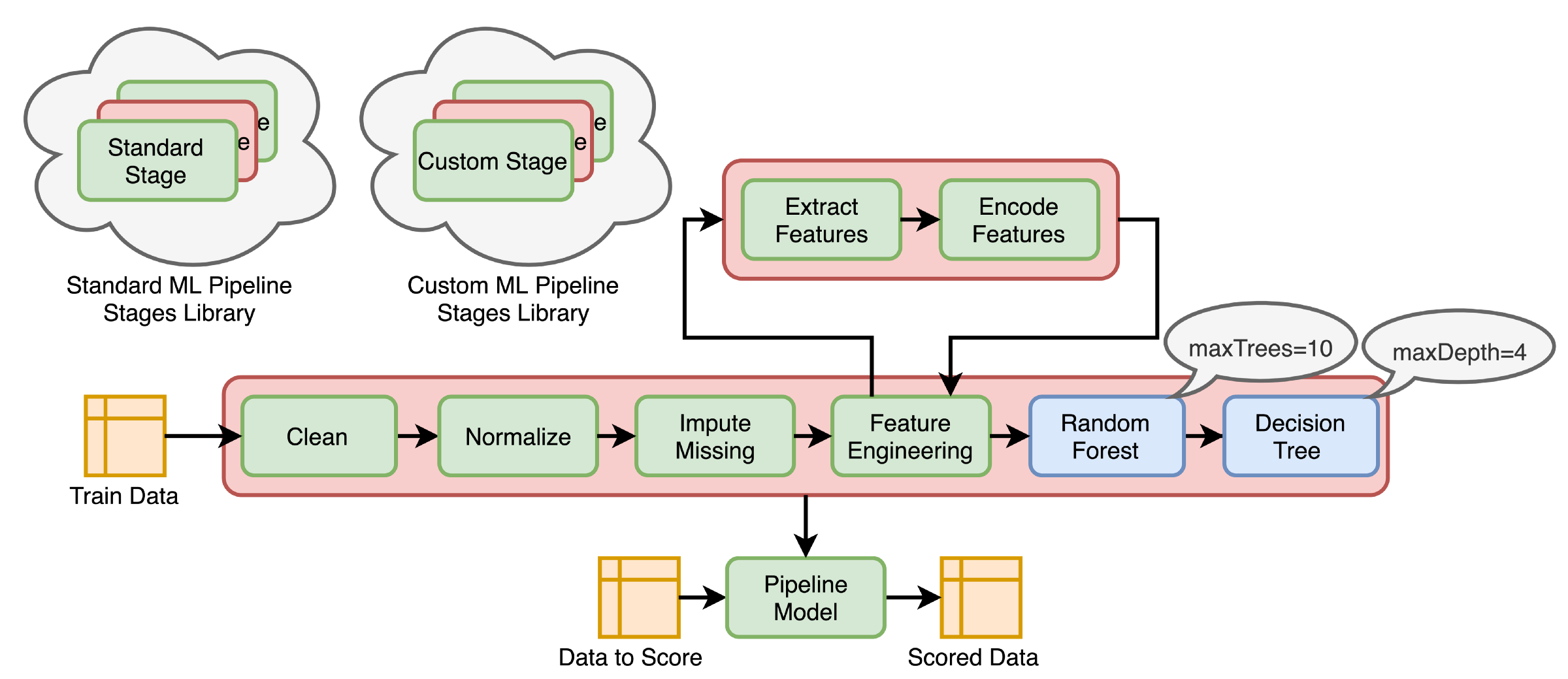
Pipelines combine multiple steps into a single workflow that can be easily run. * Data cleaning and feature processing via transformers, using stages * Model definition * Run the pipeline to do all transformations and fit the model
The Pipeline constructor takes an array of pipeline stages
Why Pipelines?
Cleaner Code: Accounting for data at each step of preprocessing can get messy. With a Pipeline, you won’t need to manually keep track of your training and validation data at each step.
Fewer Bugs: There are fewer opportunities to misapply a step or forget a preprocessing step.
Easier to Productionize: It can be surprisingly hard to transition a model from a prototype to something deployable at scale, but Pipelines can help.
More Options for Model Validation: We can easily apply cross-validation and other techniques to our Pipelines.
Example
Using the HMP Dataset. The structure of the dataset looks like this:
Let’s transform strings into numbers
- The
StringIndexeris an estimator having bothfitandtransformmethods. - To create a StringIndexer object (indexer), pass the
classcolumn as inputCol, andclassIndexas outputCol. - Then we fit the DataFrame to the indexer (running the estimator), and transform the DataFrame (running the transformer). This creates a brand new DataFrame (indexed), which we can see below, containing the classIndex additional column.
from pyspark.ml.feature import StringIndexer
indexer = StringIndexer(inputCol = 'class', outputCol = 'classIndex')
indexed = indexer.fit(df).transform(df) # This is a new data frame
# Let's see it
indexed.show(5)
+---+---+---+-----------+--------------------+----------+
| x| y| z| class| source|classIndex|
+---+---+---+-----------+--------------------+----------+
| 29| 39| 51|Drink_glass|Accelerometer-201...| 2.0|
| 29| 39| 51|Drink_glass|Accelerometer-201...| 2.0|
| 28| 38| 52|Drink_glass|Accelerometer-201...| 2.0|
| 29| 37| 51|Drink_glass|Accelerometer-201...| 2.0|
| 30| 38| 52|Drink_glass|Accelerometer-201...| 2.0|
+---+---+---+-----------+--------------------+----------+
only showing top 5 rowsOne-Hot Encode categorical variables
- Unlike the
StringIndexer,OneHotEncoderis a pure transformer, having only thetransformmethod. It uses the same syntax ofinputColandoutputCol. OneHotEncodercreates a brand new DataFrame (encoded), with acategory_veccolumn added to the previous DataFrame(indexed).OneHotEncoderdoesn’t return several columns containing only zeros and ones; it returns a sparse-vector as seen in the categoryVec column. Thus, for the ‘Drink_glass’ class above, SparkML returns a sparse-vector that says there are 13 elements, and at position 2, the class value exists(1.0).
from pyspark.ml.feature import OneHotEncoder
# The OneHotEncoder is a pure transformer object. it does not use the fit()
encoder = OneHotEncoder(inputCol = 'classIndex', outputCol = 'categoryVec')
encoded = encoder.transform(indexed) # This is a new data frame
encoded.show(5, False)
+---+---+---+-----------+----------------------------------------------------+----------+--------------+
|x |y |z |class |source |classIndex|categoryVec |
+---+---+---+-----------+----------------------------------------------------+----------+--------------+
|29 |39 |51 |Drink_glass|Accelerometer-2011-06-01-14-13-57-drink_glass-f1.txt|2.0 |(13,[2],[1.0])|
|29 |39 |51 |Drink_glass|Accelerometer-2011-06-01-14-13-57-drink_glass-f1.txt|2.0 |(13,[2],[1.0])|
|28 |38 |52 |Drink_glass|Accelerometer-2011-06-01-14-13-57-drink_glass-f1.txt|2.0 |(13,[2],[1.0])|
|29 |37 |51 |Drink_glass|Accelerometer-2011-06-01-14-13-57-drink_glass-f1.txt|2.0 |(13,[2],[1.0])|
|30 |38 |52 |Drink_glass|Accelerometer-2011-06-01-14-13-57-drink_glass-f1.txt|2.0 |(13,[2],[1.0])|
+---+---+---+-----------+----------------------------------------------------+----------+--------------+
only showing top 5 rowsAssembling the feature vector
- The
VectorAssemblerobject gets initialized with the same syntax asStringIndexerandOneHotEncoder. - The list
['x', 'y', 'z']is passed toinputCols, and we specify outputCol = ‘features’. - This is also a pure transformer.
from pyspark.ml.linalg import Vectors
from pyspark.ml.feature import VectorAssembler
# VectorAssembler creates vectors from ordinary data types for us
vectorAssembler = VectorAssembler(inputCols = ['x','y','z'], outputCol = 'features')
# Now we use the vectorAssembler object to transform our last updated dataframe
features_vectorized = vectorAssembler.transform(encoded) # note this is a new df
features_vectorized.show(5, False)
+---+---+---+-----------+----------------------------------------------------+----------+--------------+----------------+
|x |y |z |class |source |classIndex|categoryVec |features |
+---+---+---+-----------+----------------------------------------------------+----------+--------------+----------------+
|29 |39 |51 |Drink_glass|Accelerometer-2011-06-01-14-13-57-drink_glass-f1.txt|2.0 |(13,[2],[1.0])|[29.0,39.0,51.0]|
|29 |39 |51 |Drink_glass|Accelerometer-2011-06-01-14-13-57-drink_glass-f1.txt|2.0 |(13,[2],[1.0])|[29.0,39.0,51.0]|
|28 |38 |52 |Drink_glass|Accelerometer-2011-06-01-14-13-57-drink_glass-f1.txt|2.0 |(13,[2],[1.0])|[28.0,38.0,52.0]|
|29 |37 |51 |Drink_glass|Accelerometer-2011-06-01-14-13-57-drink_glass-f1.txt|2.0 |(13,[2],[1.0])|[29.0,37.0,51.0]|
|30 |38 |52 |Drink_glass|Accelerometer-2011-06-01-14-13-57-drink_glass-f1.txt|2.0 |(13,[2],[1.0])|[30.0,38.0,52.0]|
+---+---+---+-----------+----------------------------------------------------+----------+--------------+----------------+
only showing top 5 rowsNormalizing the dataset
Normalizer, like all transformers, has a consistent syntax. Looking at theNormalizerobject, it contains the parameterp=1.0(the default norm value for Pyspark Normalizer isp=2.0.)p=1.0uses Manhattan Distancep=2.0uses Euclidean Distance
from pyspark.ml.feature import Normalizer
normalizer = Normalizer(inputCol = 'features', outputCol = 'features_norm', p=1.0) # Manhattan Distance
normalized_data = normalizer.transform(features_vectorized) # New data frame too.
normalized_data.show(5)
>>
+---+---+---+-----------+--------------------+----------+--------------+----------------+--------------------+
| x| y| z| class| source|classIndex| categoryVec| features| features_norm|
+---+---+---+-----------+--------------------+----------+--------------+----------------+--------------------+
| 29| 39| 51|Drink_glass|Accelerometer-201...| 2.0|(13,[2],[1.0])|[29.0,39.0,51.0]|[0.24369747899159...|
| 29| 39| 51|Drink_glass|Accelerometer-201...| 2.0|(13,[2],[1.0])|[29.0,39.0,51.0]|[0.24369747899159...|
| 28| 38| 52|Drink_glass|Accelerometer-201...| 2.0|(13,[2],[1.0])|[28.0,38.0,52.0]|[0.23728813559322...|
| 29| 37| 51|Drink_glass|Accelerometer-201...| 2.0|(13,[2],[1.0])|[29.0,37.0,51.0]|[0.24786324786324...|
| 30| 38| 52|Drink_glass|Accelerometer-201...| 2.0|(13,[2],[1.0])|[30.0,38.0,52.0]|[0.25,0.316666666...|
+---+---+---+-----------+--------------------+----------+--------------+----------------+--------------------+
only showing top 5 rowsRunning the transformation Pipeline
The Pipeline constructor below takes an array of Pipeline stages we pass to it. Here we pass the 4 stages defined earlier, in the right sequence, one after another.
Now the Pipeline object fits to our original DataFrame df
Finally, we transform our DataFrame df using the Pipeline Object.
pipelined_data = data_model.transform(df)
pipelined_data.show(5)
+---+---+---+-----------+--------------------+----------+--------------+----------------+--------------------+
| x| y| z| class| source|classIndex| categoryVec| features| features_norm|
+---+---+---+-----------+--------------------+----------+--------------+----------------+--------------------+
| 29| 39| 51|Drink_glass|Accelerometer-201...| 2.0|(13,[2],[1.0])|[29.0,39.0,51.0]|[0.24369747899159...|
| 29| 39| 51|Drink_glass|Accelerometer-201...| 2.0|(13,[2],[1.0])|[29.0,39.0,51.0]|[0.24369747899159...|
| 28| 38| 52|Drink_glass|Accelerometer-201...| 2.0|(13,[2],[1.0])|[28.0,38.0,52.0]|[0.23728813559322...|
| 29| 37| 51|Drink_glass|Accelerometer-201...| 2.0|(13,[2],[1.0])|[29.0,37.0,51.0]|[0.24786324786324...|
| 30| 38| 52|Drink_glass|Accelerometer-201...| 2.0|(13,[2],[1.0])|[30.0,38.0,52.0]|[0.25,0.316666666...|
+---+---+---+-----------+--------------------+----------+--------------+----------------+--------------------+
only showing top 5 rowsOne last step before training
# first let's list out the columns we want to drop
cols_to_drop = ['x','y','z','class','source','classIndex','features']
# Next let's use a list comprehension with conditionals to select cols we need
selected_cols = [col for col in pipelined_data.columns if col not in cols_to_drop]
# Let's define a new train_df with only the categoryVec and features_norm cols
df_train = pipelined_data.select(selected_cols)
# Let's see our training dataframe.
df_train.show(5)
+--------------+--------------------+
| categoryVec| features_norm|
+--------------+--------------------+
|(13,[2],[1.0])|[0.24369747899159...|
|(13,[2],[1.0])|[0.24369747899159...|
|(13,[2],[1.0])|[0.23728813559322...|
|(13,[2],[1.0])|[0.24786324786324...|
|(13,[2],[1.0])|[0.25,0.316666666...|
only showing top 5 rowsYou now have a DataFrame optimized for SparkML!
Machine Learning Models in Spark
There are many Spark machine learning models
Regression
Linear regression - what is the optimization method?
Survival regression
Generalized linear model (GLM)
Random forest regression
Classification
- Logistic regression
- Gradient-boosted tree model (GBM)
- Naive Bayes
- Multilayer perception (neural network!)
Other
- Clustering (K-means, LDA, GMM)
- Association rule mining
Building your Model
Feature selection using easy R-like formulas
# Example from Spark docs
from pyspark.ml.feature import RFormula
dataset = spark.createDataFrame(
[(7, "US", 18, 1.0),
(8, "CA", 12, 0.0),
(9, "NZ", 15, 0.0)],
["id", "country", "hour", "clicked"])
formula = RFormula(
formula="clicked ~ country + hour",
featuresCol="features",
labelCol="label")
output = formula.fit(dataset).transform(dataset)
output.select("features", "label").show()What if you have too many columns for your model?
Evaluating 1,000s or 10,000s of variables?
Chi-Squared Selector
Pick categorical variables that are most dependent on the response variable
Can even check the p-values of specific variables!
# Example from Spark docs
from pyspark.ml.feature import ChiSqSelector
from pyspark.ml.linalg import Vectors
df = spark.createDataFrame([
(7, Vectors.dense([0.0, 0.0, 18.0, 1.0]), 1.0,),
(8, Vectors.dense([0.0, 1.0, 12.0, 0.0]), 0.0,),
(9, Vectors.dense([1.0, 0.0, 15.0, 0.1]), 0.0,)], ["id", "features", "clicked"])
selector = ChiSqSelector(numTopFeatures=1, featuresCol="features",
outputCol="selectedFeatures", labelCol="clicked")
result = selector.fit(df).transform(df)
print("ChiSqSelector output with top %d features selected" % selector.getNumTopFeatures())
result.show()Tuning your model
Part 1
# Example from Spark docs
from pyspark.ml.evaluation import RegressionEvaluator
from pyspark.ml.regression import LinearRegression
from pyspark.ml.tuning import ParamGridBuilder, TrainValidationSplit
# Prepare training and test data.
data = spark.read.format("libsvm")\
.load("data/mllib/sample_linear_regression_data.txt")
train, test = data.randomSplit([0.9, 0.1], seed=12345)
lr = LinearRegression(maxIter=10)
# We use a ParamGridBuilder to construct a grid of parameters to search over.
# TrainValidationSplit will try all combinations of values and determine best model using
# the evaluator.
paramGrid = ParamGridBuilder()\
.addGrid(lr.regParam, [0.1, 0.01]) \
.addGrid(lr.fitIntercept, [False, True])\
.addGrid(lr.elasticNetParam, [0.0, 0.5, 1.0])\
.build()Tuning your model
Part 2
# In this case the estimator is simply the linear regression.
# A TrainValidationSplit requires an Estimator, a set of Estimator ParamMaps, and an Evaluator.
tvs = TrainValidationSplit(estimator=lr,
estimatorParamMaps=paramGrid,
evaluator=RegressionEvaluator(),
# 80% of the data will be used for training, 20% for validation.
trainRatio=0.8)
# Run TrainValidationSplit, and choose the best set of parameters.
model = tvs.fit(train)
# Make predictions on test data. model is the model with combination of parameters
# that performed best.
model.transform(test)\
.select("features", "label", "prediction")\
.show()Project Review
Review project overview https://gu-dsan.github.io/6000-fall-2023/project/project.html
Assignment link - https://georgetown.instructure.com/courses/172712/assignments/958317
Lab time!
DSAN 6000 | Fall 2023 | https://gu-dsan.github.io/6000-fall-2023/